Prediction of Molecular Pathways and Key Mutations¶
Click to open in: [GitHub][Colab]
About this demo¶
Prediction of molecular pathways and key mutations directly from Haematoxylin and Eosin stained histology images can help bypass additional genetic (e.g., polymerase chain reaction or PCR) or immunohistochemistry (IHC) testing, which can therefore save both money and time.
In this example notebook, we show how you can use pretrained models to reproduce the inference results obtained by Bilal et al.
In this paper, an Iterative Draw and Rank Sampling (IDaRS) approach is proposed, which consists of a two-stage approach:
Patch-level tumour classification
Patch-level WSI classification
In stage 1, we use a pretrained tumour segmentation model to identify potentially diagnositc areas. In stage 2, we make a task-specific prediction for each tumour patch. For stage 2, the model is trained utilising only the slide-level label and hence the model does not require any detailed annotations at the cell or region level. Here, representative tiles from each WSI are iteratively sampled, which gives justification for the name IDaRS.
In TIAToolbox, we include models that are capable of predicting:
Microsatellite instability (MSI)
Hypermutation density
Chromosomal instability
CpG island methylator phenotype (CIMP)-high prediction
BRAF mutation
TP53 mutation
Available models¶
In line with the above description, in TIAToolbox, we provide the following pretrained models used as part of the original publication.
Tumour segmentation
resnet18-idars-tumour
Task specific prediction
MSI:
resnet34-idars-msiHypermutation density:
resnet34-idars-hmChromosomal instability:
resnet34-idars-cinCpG island methylator phenotype (CIMP)-high prediction:
resnet34-idars-cimpBRAF mutation:
resnet34-idars-brafTP53 mutation:
resnet34-idars-tp53
The provided models are trained on the first fold used in the original paper.
Downloading the required files¶
We download, over the internet, image files used for the purpose of this notebook. In particular, we download a whole slide image of cancerous colon tissue to highlight how the pipeline works.
In Colab, if you click the files icon (see below) in the vertical toolbar on the left hand side then you can see all the files which the code in this notebook can access. The data will appear here when it is downloaded.
wsi_file_name = save_dir / "sample_wsi.svs"
logger.info("Download has started. Please wait...")
# Downloading sample TCGA whole-slide image
r = requests.get(
"https://tiatoolbox.dcs.warwick.ac.uk/sample_wsis/TCGA-AD-6964-01Z-00-DX1.83AF88B9-C59B-48C6-A739-85ACB8F8ECA9.svs",
timeout=100, # 100s
)
with wsi_file_name.open("wb") as f:
f.write(r.content)
logger.info("Download is complete.")
Show code cell output
|2023-12-15|17:16:16.347| [INFO] Download has started. Please wait...
|2023-12-15|17:22:11.163| [INFO] Download is complete.
Tumour segmentation using TIAToolbox pretrained models¶
In this section, we will display patch-level tumour segmentation results using a pretrained model used in the original paper by Bilal et al. In particular, this model is a ResNet model with 18 layers (resnet18). A prediction is made for each input patch, which denotes the probability of being tumour.
More information on the model and the dataset used for training can be found here.
In line with the patch prediction model provided in tiatoolbox, the tumour segmentation model can be applied to input patches, large images tiles or whole-slide images. In order to replicate the original pipeline, we choose to process a sample whole-slide image. It can be seen that we can perform inference on a WSI with minimal effort. First we create the PatchPredictor object, which denotes the pretrained model that we will use, along with other arguments, such as the batch size and number of workers. Then, we call the predict method to process the slide and return the results. More information on using the PatchPredictor functionality can be seen in the dedicated patch prediction notebook.
wsi_file_list = [
wsi_file_name,
] # the list of WSIs to process- in this example we just use a single WSI.
tumour_predictor = PatchPredictor(
pretrained_model="resnet18-idars-tumour",
batch_size=64,
num_loader_workers=WORKERS,
)
Show code cell output
Downloading: "https://download.pytorch.org/models/resnet18-f37072fd.pth" to /root/.cache/torch/hub/checkpoints/resnet18-f37072fd.pth
100%|██████████| 44.7M/44.7M [00:00<00:00, 125MB/s]
tumour_output = tumour_predictor.predict(
imgs=wsi_file_list,
mode="wsi",
return_probabilities=True,
on_gpu=ON_GPU,
)
Show code cell output
|2023-12-15|17:22:17.014| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
100%|###########################################| 24/24 [03:05<00:00, 7.73s/it]
As can be seen above, with just a few lines of code we are able to perform tumour segmentation on whole-slide images. When running the prediction, as we did above, the default parameters for patch_shape, stride_shape and resolution will be used. Here, patch_shape, and resolution are in line with what was used for training the model, whereas stride_shape is set to be equal to the patch_shape (no overlap). In particular, the input patch size is 512x512 and the processing resolution is 0.5 microns per pixel (~20x objective magnification).
If you want to change the default parameters, you will need to define this using IOPatchPredictorConfig. For example, you may want to run with 50% overlap between neighbouring patches. Therefore, you would define:
wsi_ioconfig = IOPatchPredictorConfig(
input_resolutions=[{'units': 'mpp', 'resolution': 0.5}],
patch_input_shape=[512, 512],
stride_shape=[256, 256],
)
Then, you would add this in the predict method as follows:
tumour_output = tumour_predictor.predict(
imgs=[wsi_file_name],
mode='wsi',
return_probabilities=True,
on_gpu=True
ioconfig=wsi_ioconfig)
Below, we show how to merge the output predictions to form a 2-dimensional prediction map, denoting areas predicted as tumour. This prediction map is used in the second step of the pipeline, where we use only patches containing tumour.
# The resolution in which we desire to merge and visualize the patch predictions
overview_resolution = 1.25
# The unit of the `resolution` parameter.
# Possible values are "power", "level", "mpp", or "baseline".
overview_unit = "power"
# merge predictions to form a 2-dimensional output at the desired resolution
tumour_mask = tumour_predictor.merge_predictions(
wsi_file_name,
tumour_output[0],
resolution=overview_resolution,
units=overview_unit,
)
# the output map will contain values from 0 to 2.
# 0:background that is not processed, 1:non-tumour prediction, 2:tumour predictions
tumour_mask = tumour_mask == 2 # binarise the output # noqa: PLR2004
# let's save the tumour mask, so that we can use it in stage 2!
imwrite(str(save_dir / "tumour_mask.png"), tumour_mask.astype("uint8") * 255)
|2023-12-15|17:25:23.464| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
Now that we have merged the prediction, let’s visualise the results!
# first read the WSI at a low-resolution.
# Here, we use the same resolution that was used when merging the patch-level results.
wsi = WSIReader.open(wsi_file_name)
wsi_overview = wsi.slide_thumbnail(resolution=overview_resolution, units=overview_unit)
# [Overlay map creation]
# Creating label-color dictionary to be fed into `overlay_prediction_mask` function
# This helps to generate a color legend.
label_dict = {"Non-Tumour": 0, "Tumour": 1}
label_color_dict = {}
colors = [[255, 255, 255], [255, 0, 0]] # defining colours for overlay (white and red)
for class_name, label in label_dict.items():
label_color_dict[label] = (class_name, np.array(colors[label]))
overlay = overlay_prediction_mask(
wsi_overview,
tumour_mask,
alpha=0.5,
label_info=label_color_dict,
return_ax=True,
)
plt.show()
|2023-12-15|17:25:39.212| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
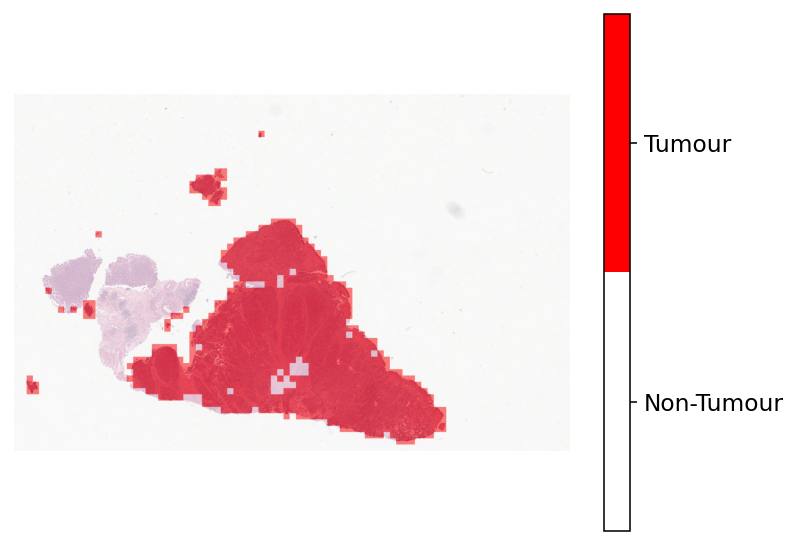
WSI prediction using TIAToolbox pretrained models¶
Next, we show how one can use a second CNN that takes as input the results obtained from part 1 and gives a prediction for each tumour patch in the input WSI. In the original paper, 4 fold cross validation was used. In the toolbox, we choose to provide models trained on the first fold of the data, which enables us to cross check the results and ensure that they are in line with the original work. All models in the second step of the pipeline are ResNet34.
To speed up this step, we choose to retrain the models without stain normalization (as oppoed to the original IDaRS) and instead with colour jitter. We report the difference in results at the end of this notebook.
Near the beginning of the notebook, we mention which prediction tasks we consider. Next, we will run inference for all of these prediction tasks and visualise the output.
# Run inference for each of the 6 WSI prediction tasks
prediction_tasks = ["msi", "braf", "cimp", "cin", "hm", "tp53"]
# iterate over each of the prediction tasks and add the results to a dictionary
wsi_output_dict = {}
for task in prediction_tasks:
wsi_predictor = PatchPredictor(
pretrained_model=f"resnet34-idars-{task}",
batch_size=64,
num_loader_workers=WORKERS,
)
# Obtained tumour mask from stage 1 to only process patches from those regions.
wsi_output_dict[task] = wsi_predictor.predict(
imgs=wsi_file_list,
masks=[save_dir / "tumour_mask.png"],
mode="wsi",
return_probabilities=True,
on_gpu=ON_GPU,
)
Show code cell output
Downloading: "https://download.pytorch.org/models/resnet34-b627a593.pth" to /root/.cache/torch/hub/checkpoints/resnet34-b627a593.pth
100%|██████████| 83.3M/83.3M [00:00<00:00, 146MB/s]
100%|###########################################| 22/22 [02:10<00:00, 5.95s/it]
100%|###########################################| 22/22 [02:08<00:00, 5.84s/it]
100%|###########################################| 22/22 [02:05<00:00, 5.70s/it]
100%|###########################################| 22/22 [02:08<00:00, 5.86s/it]
100%|###########################################| 22/22 [02:01<00:00, 5.52s/it]
100%|###########################################| 22/22 [02:07<00:00, 5.80s/it]
Now that we have performed patch-level classification for each tumour patch, let’s merge together the results to get the 2D prediction map. Here, we return the raw probability map, rather than the class predictions. We do this because the final step in the IDaRS pipeline uses the raw patch-level probabilities to classify each WSI.
# The resolution in which we desire to merge and visualize the patch predictions
overview_resolution = 1.25
# The unit of the `resolution` parameter.
# Possible values are "power", "level", "mpp", or "baseline"
overview_unit = "power"
# merge predictions to form a 2-dimensional output at the desired resolution
merged_output_dict = {}
for task in prediction_tasks:
merged_output = PatchPredictor.merge_predictions(
wsi_file_name,
wsi_output_dict[task][0],
resolution=overview_resolution,
units=overview_unit,
return_raw=True,
)
merged_output_dict[task] = merged_output[..., 1] # consider only the positive class
|2023-12-15|17:47:53.205| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
|2023-12-15|17:47:53.408| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
|2023-12-15|17:47:53.595| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
|2023-12-15|17:47:53.794| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
|2023-12-15|17:47:53.980| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
|2023-12-15|17:47:54.189| [WARNING] Read: Scale > 1.This means that the desired resolution is higher than the WSI baseline (maximum encoded resolution). Interpolation of read regions may occur.
Now that we have merged the predictions to obtain the probability map, let’s visualise the results. This is very similar to the overlay_patch_prediction function, but label_info does not need to be provided because we only have one class. Also, min_val can be provided. This is a number between 0 and 1 which ensures that the probability map is only shown for values greater than min_val.
# [Overlay map creation]
# iterate over the tasks and generate the probability map
overlay_list = []
for task in prediction_tasks:
# only show the probability map when it is greater than `min_val` (0.1 is used here)
overlay_prob_map = overlay_probability_map(
wsi_overview,
merged_output_dict[task],
alpha=0.5,
min_val=0.1,
return_ax=False,
)
overlay_list.append(overlay_prob_map)
# visualise the overlays in a single plot
fig = plt.figure(figsize=(14, 7))
for i in range(6):
ax = plt.subplot(2, 3, i + 1)
plt.imshow(overlay_list[i])
plt.axis("off")
plt.title(prediction_tasks[i].upper())
fig.suptitle("Probability Map Overlay for Each Task")
plt.show()
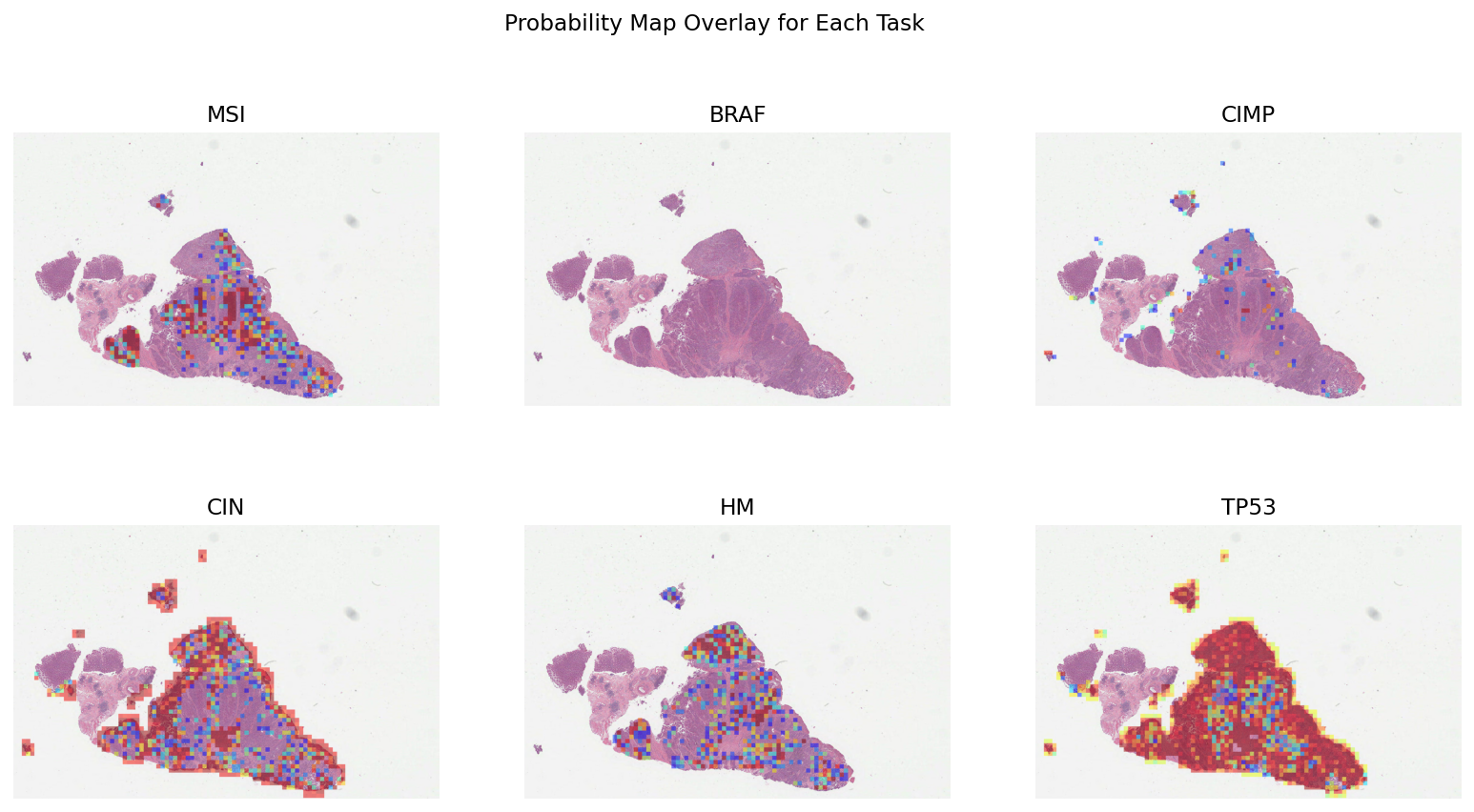
These probability maps can be used to increase the interpretability of results and help identify regions contributing to the overall prediction. To get a smoother output, you can increase the overlap by modifying IOPatchPredictorConfig.
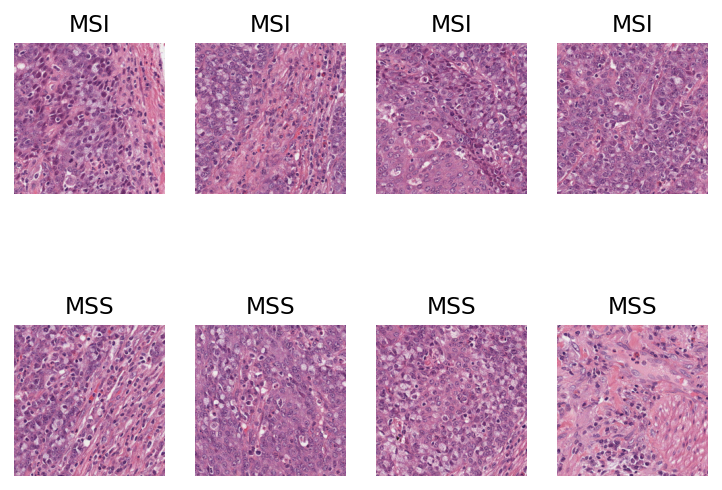
Let’s visualise some patches that have either a high probability or a low probability of being MSI positive. For this, we randomly sample 4 patches with probability of being MSI > 0.95 and 4 patches with probability of being MSI < 0.05.
# get the probabilities of each processed patch being MSI
msi_probabilities = wsi_output_dict["msi"][0]["probabilities"]
msi_probabilities = np.array(msi_probabilities)[..., 1]
# get the coordinates of each processed patch
msi_coordinates = wsi_output_dict["msi"][0]["coordinates"]
msi_coordinates = np.array(msi_coordinates)
# subset where MSI probability is greater than 0.95
threshold = 0.95
msi_probabilities_subset_pos = msi_probabilities[msi_probabilities > threshold]
msi_coordinates_subset_pos = msi_coordinates[msi_probabilities > threshold]
# subset where MSI probability is less than than 0.05
threshold = 0.05
msi_probabilities_subset_neg = msi_probabilities[msi_probabilities > threshold]
msi_coordinates_subset_neg = msi_coordinates[msi_probabilities > threshold]
msi_coordinates_subset = list(msi_coordinates_subset_pos) + list(
msi_coordinates_subset_neg,
)
# randomly sample 4 positive and 4 negative
rng = np.random.default_rng() # Numpy Random Generator
random_pos_idx = rng.integers(0, msi_probabilities_subset_pos.shape[0], size=4)
random_neg_idx = rng.integers(0, msi_probabilities_subset_neg.shape[0], size=4)
random_idx = list(random_pos_idx) + list(random_neg_idx)
Now that we have obtained our random sample, let’s plot the patches! First, we use TIAToolbox’s WSIReader to read the original WSI and extract patches at defined locations. For detailed information on this, see the relevant example notebook!
# read the WSI and get the resolution used during processing.
wsi_reader = WSIReader.open(input_img=wsi_file_name)
resolution = wsi_output_dict["msi"][0]["resolution"]
units = wsi_output_dict["msi"][0]["units"]
# visualise the overlays in a single plot
fig = plt.figure()
for i in range(8):
ax = plt.subplot(2, 4, i + 1)
coords = msi_coordinates_subset[random_idx[i]]
size = (
coords[2] - coords[0],
coords[3] - coords[1],
) # determine the size of patch from the coordinates
# get the patch
patch = wsi_reader.read_rect(
(coords[0] * 4, coords[1] * 4),
size,
resolution=resolution,
units=units,
)
plt.imshow(patch)
plt.axis("off")
if i < 4: # noqa: PLR2004
plt.title("MSI")
else:
plt.title("MSS")
plt.show()
The probability maps are not directly needed to obtain the WSI-level score. Below, we will demonstrate how one can obtain the WSI score utilising the output from the PatchPredictor in step 2. For the purpose of this example, we will show how to use mean and max aggregation to obtain a slide-level prediction score.
First, let’s examine the format of the dictionary output by PatchPredictor. We look only at the MSI output as an example, but the output from each task is similar.
# get the keys of the dictionary returned at the output of the MSI CNNPatchPredictor
patch_pred_keys = list(wsi_output_dict["msi"][0].keys())
logger.info(
"Output of PatchPredictor: [%s, %s, %s, %s, %s, %s, %s.]",
patch_pred_keys[0],
patch_pred_keys[1],
patch_pred_keys[2],
patch_pred_keys[3],
patch_pred_keys[4],
patch_pred_keys[5],
patch_pred_keys[6],
)
|2023-12-15|17:48:04.998| [INFO] Output of PatchPredictor: [probabilities, predictions, coordinates, label, pretrained_model, resolution, units.]
As we can see, the probabilities are returned as output. Now, let’s compute, for each task, the average and maximum over all probabilities of tumour tiles being positive.
slide_score_mean = {}
slide_score_max = {}
# iterate over the tasks
for task in prediction_tasks:
probabilities = np.array(wsi_output_dict[task][0]["probabilities"])[
...,
1,
] # only consider positive class
slide_score_mean[task] = np.mean(
probabilities,
) # get the average probability over all tumour tiles
slide_score_max[task] = np.max(
probabilities,
) # get the maximum probability over all tumour tiles
# print the scores
print("MEAN AGGREGATION") # noqa: T201
for task, value in slide_score_mean.items():
print(task, ":", value) # noqa: T201
print("-" * 30) # noqa: T201
print("MAX AGGREGATION") # noqa: T201
for task, value in slide_score_max.items():
print(task, ":", value) # noqa: T201
MEAN AGGREGATION
msi : 0.19757118561392417
braf : 0.001280215530521352
cimp : 0.04703372328468129
cin : 0.4985422825156989
hm : 0.20540336819709304
tp53 : 0.8004594413849939
------------------------------
MAX AGGREGATION
msi : 0.9999943971633911
braf : 0.04610889405012131
cimp : 0.997495174407959
cin : 0.9994856119155884
hm : 0.9999963045120239
tp53 : 0.9997155070304871
As can be seen, it is straight forward to go from the WSI output to the WSI-level prediction score. In a similar way, different aggregation methods can be used, such as top k probabilities. To highlight how easy it is to obtain the slide level score using TIAToolbox, we provide below the code required to go from input to output.
# TUMOUR DETECTION
tumour_predictor = PatchPredictor(pretrained_model='resnet18-idars-tumour', batch_size=64)
tumour_output = tumour_predictor.predict(
imgs=[wsi_file_name],
mode='wsi',
return_probabilities=True,
on_gpu=True)
tumour_mask = tumour_predictor.merge_predictions(
wsi_file_name,
tumour_output[0],
resolution=overview_resolution,
units=overview_unit)
tumour_mask = tumour_mask == 2 # binarise the output
imwrite(save_dir + 'tumour_mask.png',
tumour_mask.astype('uint8') * 255)
# WSI PREDICTION
msi_predictor = PatchPredictor(pretrained_model='resnet34-idars-msi', batch_size=64)
msi_output = msi_predictor.predict(
imgs=[wsi_file_name],
masks=[save_dir + 'tumour_mask.png'],
mode='wsi',
return_probabilities=True,
on_gpu=True)
# SLIDE-LEVEL SCORE
msi_probabilities = np.array(msi_output[0]['probabilities'])[...,1] # only consider MSI class
average_msi_probability = np.mean(msi_probabilities) # get the average over all tumour tiles
We encourage you to play around with the models provided by TIAToolbox for prediction of molecular pathways and key mutations. To make a different WSI level prediction, all you need to do is use a different task-specific pretrained weights in the second PatchPredictor.
Below, we report the results obtained with our retrained model compared to the original results. For the purpose of this, we only consider mean aggregation for each task. All results show the area under the receiver operating characteristic curve (AUROC).
| Task | Original | TIAToolbox | | —- | ——– | ———- | | MSI | 0.83 | 0.87 | | TP53 | 0.76 | 0.75 | | BRAF | 0.81 | 0.75 | | CIMP | 0.85 | 0.75 | | CIN | 0.85 | 0.81 | | HM | 0.86 | 0.79 |